G-quadruplexes
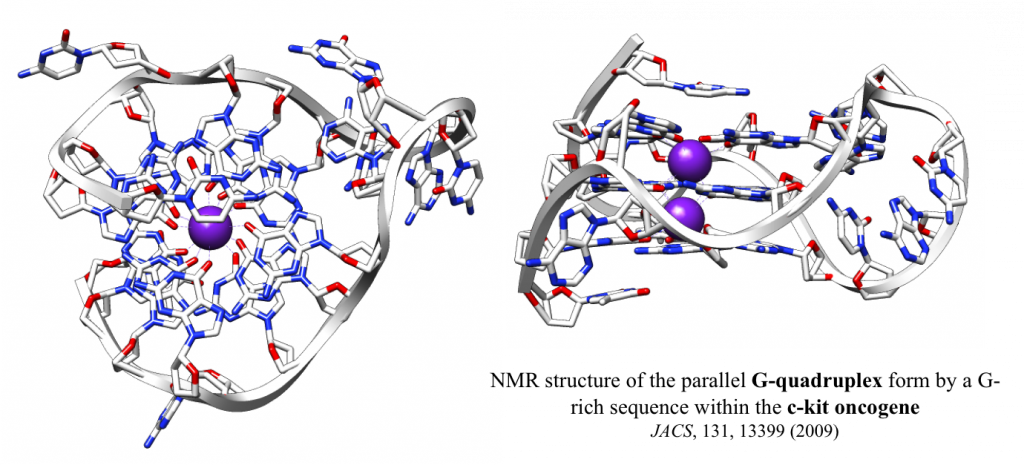
Nucleic acids are highly flexible molecules than can adopt different conformational structures. While in living systems DNA is largely double helical and RNA is single stranded, guanine-rich sequences can exist in alternative structural forms known as G-quadruplex nucleic acids. We are investigating the functional relevance of a quadruple helical form of nucleic acids and its implication for the biology of nucleic acids. The G-quadruplex hypothesis is of fundamental importance to life and may well hold the key to new therapeutic approaches in numerous areas of human disease that include cancer.
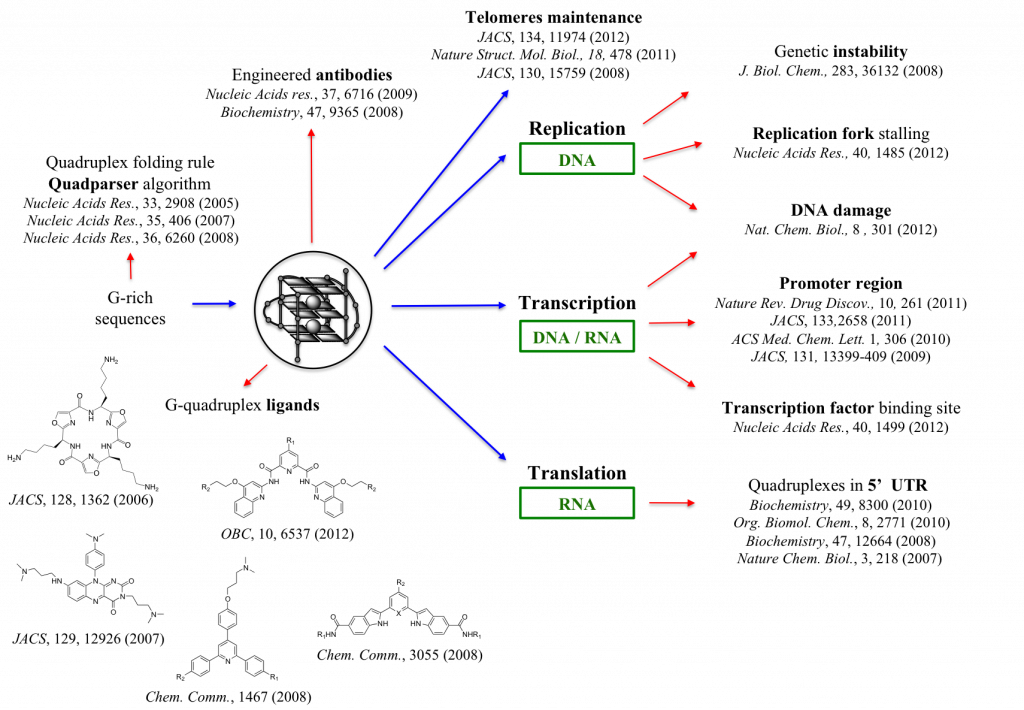
Specific mechanisms under investigation include: DNA G-quadruplex formation at the telomeres and their importance for genomic stability and replication; DNA G-quadruplexes in gene promoters and the regulation of transcription; RNA G-quadruplexes in the untranslated regions of mRNA and the control of protein synthesis (translation).
As part of our studies, we are synthesising new molecules that stabilise the nucleic acid quadruple helix and interfere with specific cellular processes. We make extensive use of biophysical methods (NMR, UV, CD and fluorescence spectroscopy) to study quadruplexes and their interactions with molecules. We are also employing computational methods (bioinformatics) and genomics to explore quadruplexes in genomes.
Key Papers
Sequencing DNA methylation and hydroxymethylation at co-occurring chromatin features
R de Cesaris Araujo Tavares, S Dhir, X He, J Monahan, M Taipale, P Golder, A Ciau-Uitz, W Gosal, D Tannahill and S Balasubramanian
Nature Communications, 2026, in press
DOI:10.1038/s41467-026-69429-6
5-methylcytosine and 5-hydroxymethylcytosine are synergistic biomarkers for early detection of colorectal cancer
F Puddu, A Johansson, A Modat, J Scotcher, R Sethi, S Yu, N Harding, M Hill, E Lleshi, C Lumby, J Tayssandier, M Wilson, R Crawford, T Charlesworth, P Creed, S Balasubramanian and R J Osborne
Communications Medicine, 2026, 6:12
DOI:10.1038/s43856-025-01278-8
Commentary on ‘Untargeted CUT&Tag reads are enriched at accessible chromatin and restrict identification of potential G4-forming sequences in G4-targeted CUT&Tag experiments’
L Melidis, R de Cesaris Araujo Tavares, X He, S Dhir, D Tannahill and S Balasubramanian
Nucleic Acids Research, 2025, 53:22
DOI:10.1093/nar/gkaf1337
An unnatural base pair for the detection of epigenetic cytosine modifications in DNA
D Schmidl, S M Becker, J M Edgerton and S Balasubramanian
Nat. Chem., 2025, 17, 1732–1741
DOI: 10.1038/s41557-025-01925-6
SCoTCH-seq reveals that 5-hydroxymethylcytosine encodes regulatory information across DNA strands
J S Hardwick, S Dhir, A Kirchner, A Simeone, S M Flynn, J M Edgerton, R de Cesaris Araujo Tavares, I Esain-Garcia, D Tannahill, P Golder, J M Monahan, W S Gosal and S Balasubramanian
PNAS, 2025, 122 (31), e2512204122
DOI: 10.1073/pnas.2512204122
DNA G-quadruplex structures act as functional elements in ⍺- and ß-globin enhancers
C Doyle, K Herka, S M Flynn, L Melidis, S Dhir, S Schoenfelder, D Tannahill and S Balasubramanian
Genome Biology, 2025, 26:155
DOI: 10.1186/s13059-025-03627-1